UNITED STATES
SECURITIES AND EXCHANGE COMMISSION
Washington, D.C. 20549
FORM 8-K
CURRENT REPORT
Pursuant to Section 13 or 15(d) of the Securities Exchange Act of 1934
Date of Report (Date of earliest event reported): January 12, 2021
BEAM THERAPEUTICS INC.
(Exact name of registrant as specified in its charter)
|
Delaware |
|
001-39208 |
|
81-5238376 |
|
(State or other jurisdiction |
|
(Commission |
|
(IRS Employer |
|
of incorporation) |
|
File Number) |
|
Identification No.) |
|
|
|
|
|
|
|
26 Landsdowne St. |
|
|
|
|
|
Cambridge, MA |
|
|
|
02139 |
|
(Address of principal executive offices) |
|
|
|
(Zip Code) |
(Registrant’s telephone number, including area code): (857) 327-8775
Not Applicable
(Former name or former address, if changed since last report)
Check the appropriate box below if the Form 8-K filing is intended to simultaneously satisfy the filing obligation of the registrant under any of the following provisions (see General Instruction A.2. below):
|
☐ |
Written communications pursuant to Rule 425 under the Securities Act (17 CFR 230.425) |
|
☐ |
Soliciting material pursuant to Rule 14a-12 under the Exchange Act (17 CFR 240.14a-12) |
|
☐ |
Pre-commencement communications pursuant to Rule 14d-2(b) under the Exchange Act (17 CFR 240.14d-2(b)) |
|
☐ |
Pre-commencement communications pursuant to Rule 13e-4(c) under the Exchange Act (17 CFR 240.13e-4(c)) |
Securities registered pursuant to Section 12(b) of the Act:
|
Title of each class |
Trading Symbol(s) |
Name of each exchange on which registered |
|
Common Stock, par value $0.01 per share |
BEAM |
Nasdaq Global Select Market |
Indicate by check mark whether the registrant is an emerging growth company as defined in Rule 405 of the Securities Act of 1933 (§230.405 of this chapter) or Rule 12b-2 of the Securities Exchange Act of 1934 (§240.12b-2 of this chapter).
Emerging growth company ☒
If an emerging growth company, indicate by check mark if the registrant has elected not to use the extended transition period for complying with any new or revised financial accounting standards provided pursuant to Section 13(a) of the Exchange Act. ☐
Item 7.01 Regulation FD Disclosure.
Beam Therapeutics Inc. (the “Company”) will be conducting meetings with participants attending the 39th Annual J.P. Morgan Healthcare Conference (the “Conference”). The slides to be presented by the Company at the Conference are furnished with this report as Exhibit 99.1, which is incorporated herein by reference.
The information in this Form 8-K (including Exhibit 99.1 attached hereto) is being furnished and shall not be deemed “filed” for purposes of Section 18 of the Securities Exchange Act of 1934, as amended (the “Exchange Act”), or otherwise subject to the liabilities of that section, nor shall it be deemed incorporated by reference into any filing by the Company, under the Securities Act of 1933, as amended, or the Exchange Act, except as expressly set forth by specific reference in such filling.
Item 9.01 Financial Statements and Exhibits.
(d) Exhibits
|
Exhibit No. |
|
Description |
|
|
|
|
|
99.1 |
|
Beam Therapeutics Inc. Presentation at the 39th Annual J.P. Morgan Healthcare Conference |
SIGNATURES
Pursuant to the requirements of the Securities Exchange Act of 1934, the registrant has duly caused this report to be signed on its behalf by the undersigned hereunto duly authorized.
|
|
|
BEAM THERAPEUTICS INC. |
|
|
|
|
|
|
|
Date: January 12, 2021 |
|
By: |
/s/ John Evans |
|
|
|
|
John Evans |
|
|
|
|
Chief Executive Officer |

PRECISION GENETIC MEDICINES THROUGH BASE EDITING Beam Therapeutics NASDAQ: BEAM Exhibit 99.1

This presentation contains forward-looking statements within the meaning of the Private Securities Litigation Reform Act of 1995. Such forward-looking statements reflect, among other things, our current expectations and anticipated results of operations, all of which are subject to known and unknown risks, uncertainties and other factors that may cause our actual results, performance or achievements, market trends, or industry results to differ materially from those expressed or implied by such forward-looking statements. Therefore, any statements contained herein that are not statements of historical fact may be forward-looking statements and should be evaluated as such. Without limiting the foregoing, the words “anticipate,” “expect,” “suggest,” “plan,” “believe,” “intend,” “project,” “forecast,” “estimates,” “targets,” “projections,” “should,” “could,” “would,” “may,” “might,” “will,” and the negative thereof and similar words and expressions are intended to identify forward-looking statements. Each forward-looking statement is subject to risks and uncertainties that could cause actual results to differ materially from those expressed or implied in such statement, including, without limitation, risks and uncertainties related to: our ability to develop, obtain regulatory approval for, and commercialize our product candidates, which may take longer or cost more than planned; our ability to raise additional funding, which may not be available; our ability to obtain, maintain and enforce patent and other intellectual property protection for our product candidates; the potential impact of the COVID-19 pandemic; that preclinical testing of our product candidates and preliminary or interim data from preclinical and clinical trials may not be predictive of the results or success of ongoing or later clinical trials; that enrollment of our clinical trials may take longer than expected; that our product candidates may experience manufacturing or supply interruptions or failures; risks related to competitive products; and the other risks and uncertainties identified under the heading “Risk Factors” and elsewhere in our annual report on Form 10-K for the year ended December 31, 2019, our quarterly reports on Form 10-Q for the quarters ended March 31, 2020, June 30, 2020 and September 30, 2020, and in any subsequent filings with the Securities and Exchange Commission (the “SEC”) which are available on the SEC’s website at www.sec.gov. Additional information will be made available by our annual and quarterly reports and other filings that we make from time to time with the SEC. These forward-looking statements (except as otherwise noted) speak only as of the date of this presentation. Factors or events that could cause our actual results to differ may emerge from time to time, and it is not possible for us to predict all of them. We undertake no obligation to update any forward-looking statement, whether as a result of new information, future developments or otherwise, except as may be required by applicable law. Forward-looking statements . 2

Coming era of one-time, curative therapies Gene editing for rare and common diseases Platform for rapidly-programmable precision medicines Our vision is to provide life-long cures for patients suffering from serious diseases 3

Base editing is a new approach to gene editing Nuclease editing CRISPR, Zinc Fingers, TALEs Base editing 4

Base editing is a highly-differentiated, potentially best-in-class gene editing technology

Diversified portfolio of wholly-owned base editing programs LNP = Lipid Nanoparticle; AAV = Adeno Associated Virus; CNS = Central Nervous System 6

Key progress and milestones 7 ELECTRO- PORATION NON-VIRAL (LNP) ✓ ✓ 2020 achievements VIRAL (AAV) ✓ ✓ In vivo proof of concept and development candidate nomination for BEAM-101 Initiate IND-enabling studies for BEAM-101 >80% direct correction of sickle mutation and development candidate nomination for BEAM-102 In vivo proof of concept and development candidate nomination for BEAM-201 In vivo proof of concept for direct correction of alpha-1 antitrypsin deficiency Research collaboration with Institute Ophthalmology Basel for preclinical development of Stargardt program ✓ Next milestones in 2021 File IND for BEAM-101 during the second half Evaluate Beam LNP formulation in non-human primates and release initial data in early 2021 Nominate first liver development candidate Initiate IND-enabling studies for BEAM-102 ✓ In vivo editing of both R83C and Q347X mutations in GSDIa mouse models ✓ Initiate IND-enabling studies for BEAM-201 Initiate non-human primate studies for Stargardt

We are establishing the leading platform for precision genetic medicine Full suite of gene editing technologies Base editing Nuclease editing RNA editing Prime editing Full suite of delivery technologies Auto / allo cell products mRNA LNP vectors Viral vectors Wholly-owned manufacturing capability 100,000 square foot GMP clinical/commercial facility in NC, phased build, online by 2023 8

Electroporation Ex vivo delivery of base editors using clinically validated electroporation technology Target cell Electroporation of program-specific gRNA and base editor mRNA Base editor performs gRNA-dependent editing 9

Autologous ex vivo cell process for editing hematopoietic stem cells Collect cells Electroporate Infuse cell product Condition patient Sickle Cell Disease Patient Sickle Cell Disease Patient 3 4 1 2 10

Sickle cell disease (SCD) T-to-A mutation causes sickling Adult β-globin gene β-globin gene Glutamic acid at 6th amino acid (HbA) Normal red blood cells Sickle β-globin gene Valine at 6th amino acid (HbS) Sickling red blood cells 11 Approximately 100,000 sickle cell disease patients in the US

BEAM-101: Recreating hereditary persistence of fetal hemoglobin (HPFH) with base editing Reactivating fetal hemoglobin genes Sickle cell disease patient Naturally-occurring base changes cause Hereditary Persistence of Fetal Hemoglobin (HPFH), which protects patients from SCD/B-Thal Base editors can reproduce these changes, leading to elevated levels of fetal hemoglobin Higher fetal hemoglobin likely to correlate with further reductions in disease symptoms Mussallam, et al. 2012. Blood. Adult sickle β-globin (HbS) Fetal γ-globin (HbF) Fetal γ-globin (HbF) HBB HBG1 HBG2 X X ABE edits regulatory element of both fetal hemoglobin genes, without cutting DNA 12
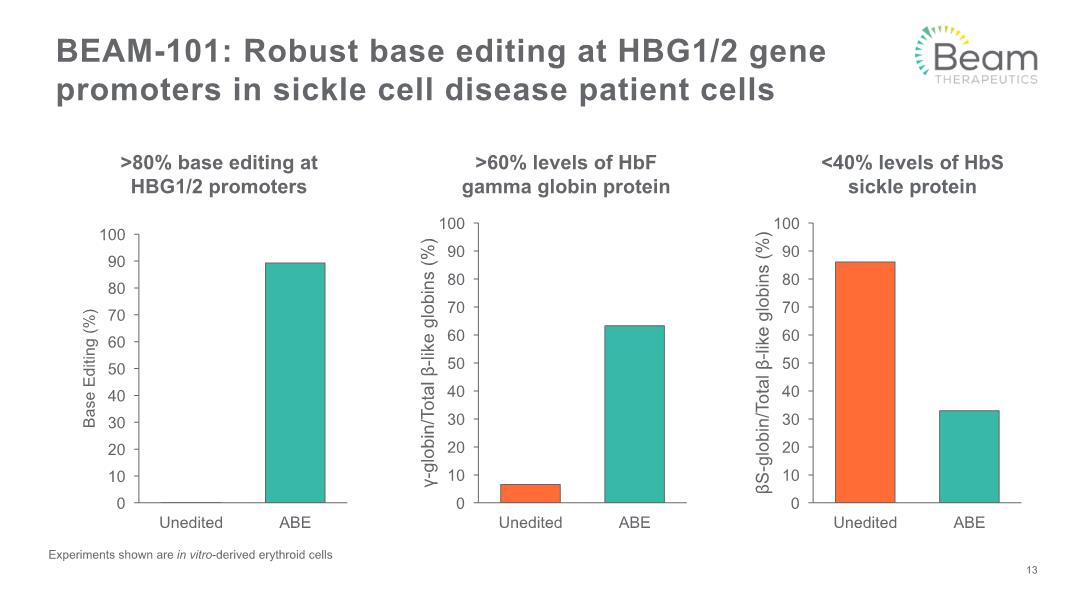
BEAM-101: Robust base editing at HBG1/2 gene promoters in sickle cell disease patient cells 13 Experiments shown are in vitro-derived erythroid cells
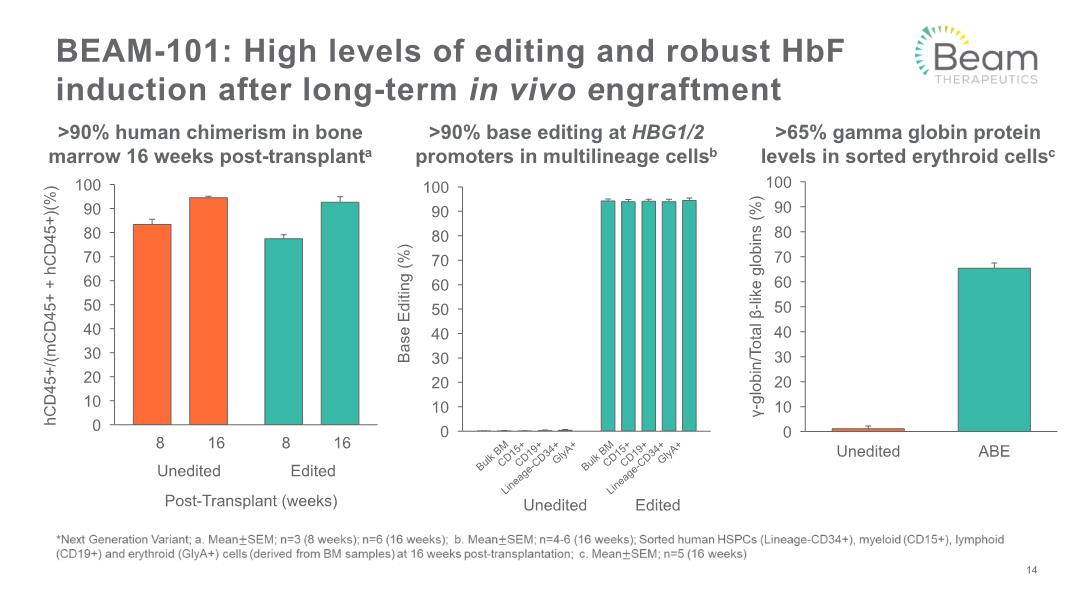
BEAM-101: High levels of editing and robust HbF induction after long-term in vivo engraftment >90% human chimerism in bone marrow 16 weeks post-transplanta >90% base editing at HBG1/2 promoters in multilineage cellsb >65% gamma globin protein levels in sorted erythroid cellsc 14 Unedited Edited Bulk BM CD15+ CD19+ Lineage-CD34+ GlyA+ Bulk BM CD15+ CD19+ Lineage-CD34+ GlyA+

BEAM-102: Direct correction of the sickle causing mutation Base editing recreates naturally-occurring human variant Hb-G Makassar which has alanine (E6A) instead of sickle-causing valine (E6V)1 Hb-G Makassar is a normal β-globin variant and does not cause sickle disease, e.g., blood smear shows negative for sickle cells2 1. Cummings, MR. 2006. Human Heredity: Principles and Issues, Updated Edition.; 2. Mohamad, et al. 2018. Hematology Reports. Common β-globin (Normal) Glutamic acid (HbA) Valine (HbS) Sickle β-globin (Disease) Makassar β-globin (Normal) Alanine (HbG) 15

BEAM-102: Highly efficient, novel direct correction of sickle mutation in sickle patient cells Allelic editing of 100 individual erythroid colonies post-electroporation was assessed for the Makassar edit by NGS using cells from HbSS donor. ~93% of cells have at least one sickle allele converted to Makassar and are potentially cured 73% Biallelic Makassar Edited Cells 20% Monoallelic Makassar Edited Cells 4% Monoallelic Makassar + Bystander Edit 2% Unedited 80% Sickle Makassar correction in sickle patient CD34 cells with ABE (N=100) 16

BEAM-102: High editing of sickle mutation leads to significant elimination of HbS globin in patient donor cells Sickle (HbS) and Makassar variant globin protein, at varying bulk levels of Makassar editing assessed by NGS, was measured by UPLC and expressed as a fraction of total beta globin in 18 day mature RBCs derived from edited HbSS CD34+s. UPLC was conducted on n = 2 for each bulk editing condition. CD34+ HbSS cells were edited and subsequently differentiated to generate mature erythroid red blood cells and exposed to low oxygen conditions (<2%) in a hypoxic chamber. Image is representative of n=2 different sickling assays from n=2 independent donors that were successfully edited at high levels (>80% by NGS) and confirmed to have near 90% Makassar globin by UPLC. 17 Sickling in unedited HbSS cells ~89% Hb G-Makassar by UPLC Elimination of HbS globin

Two base editing approaches to silence genes Expanding list of targets for CAR-T editing1 Two strategies for silencing with base editors Splice disruption with CBE CONFIDENTIAL 18 1. Adapted from Jung & Lee. 2018. Molecules and Cells.

Allogeneic multiplex edited CAR-T cell process Collect T-cells Electroporate and transduce Lymphodeplete and infuse Expand and freeze Editor mRNA gRNA #1 gRNA #2 gRNA #3 gRNA #4 Lentiviral CAR 3 4 1 2 19

BEAM-201: Multiplex editing across four genomic loci with base editing 20 Single electroporation of base editor mRNA and 4 gRNAs With few options to induce a second remission, post-relapse survival for T-cell acute lymphoblastic leukemia (T-ALL) is low TRAC: Prevent graft-vs-host disease CD52: Enable an allogeneic cell source to reduce host rejection of BEAM-201 cells PDCD1: Minimize immunosuppression of BEAM-201 cells and prolong efficacy for attacking the tumor CD7: Prevent fratricide (i.e., CAR-T cells attacking each other before they can attack the tumor)

Clinical scale BEAM-201 process produces 96-99% individual gene knock-out & potent in vivo activity 21 Clinical-scale process produces individually 96-99% editing, estimated 91% quad edited Potent in vivo tumor clearance or control across a 25-fold dose Treatment Randomization

Multiplex base editing with no detectable translocations and maintained cell expansion 22 1. Lower limit of detection for rearrangements of 0.1%; 2. B2M-B only measurable if translocation includes local rearrangement in B2M; 3. Extensive guide screen across three targets, with both BE4 and spCas9 sgRNAs, selected for high editing efficiency and expansion in single-plex test, final cell yields compared between 1, 2 and 3 edits using BE4 and spCas9, normalized to electroporation only control; B2M = beta 2 microglobulin gene; TRAC = T-cell receptor alpha constant chain gene; PDCD1 = programmed cell death protein 1 gene; Unlike Cas9, no detectable CBE-induced rearrangements in triple-edited T cells (3 donors)1 Higher yield for CBE triple-edited cells relative to Cas9 triple-edited cells (N=6)3
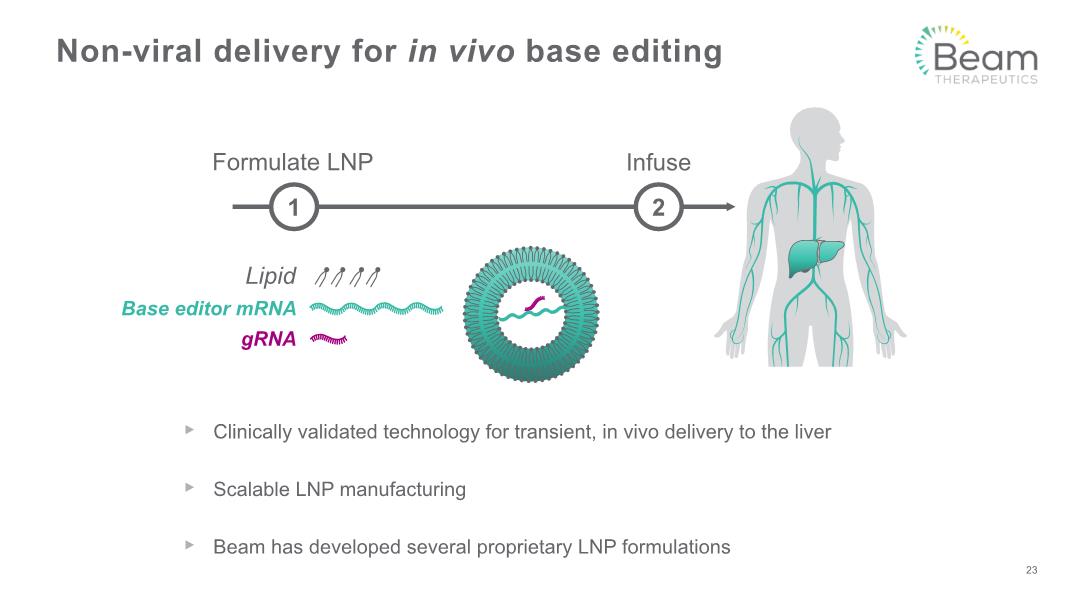
Clinically validated technology for transient, in vivo delivery to the liver Scalable LNP manufacturing Beam has developed several proprietary LNP formulations Non-viral delivery for in vivo base editing 23 Infuse Formulate LNP 1 2

Alpha-1 Antitrypsin (A1AT) Deficiency Normal A1AT Function Genetics Liver Respiratory A1AT Deficiency Wild type SERPINA1 gene A1AT protein is secreted, protecting lungs E342K (PiZ) mutation Direct correction of the PiZ mutation through base editing can: Restore circulation of functional A1AT to protect the lungs Reduce liver toxicity caused by mutant protein aggregates 24 A1AT aggregates and causes liver damage/failure Low A1AT in circulation causing lung damage, emphysema, etc

Direct correction of PiZ mutation by base editing in primary PiZZ fibroblasts Alpha-1 Antitrypsin Deficiency base editing products ~45% correction of PiZ mutation in primary PiZZ fibroblasts ABE edits Resulting A1AT Variant Target Target + Bystander Bystander only Beneficial alleles are secreted and have elastase activity 25 WT WT + D341G WT WT + D341G E342K + D341G SERPINA1 (PiZ) gene sequence

PiZ correction may confer a proliferative advantage to edited hepatocytes LNP-mediated in vivo correction of the PiZ mutation increases over time 26 P value assessed by T-test

In vivo direct correction of A1AT mutation with base editing – addressing both liver and lung 27 4.9-fold increase in functional A1AT secretion Reduction in toxic liver aggregates
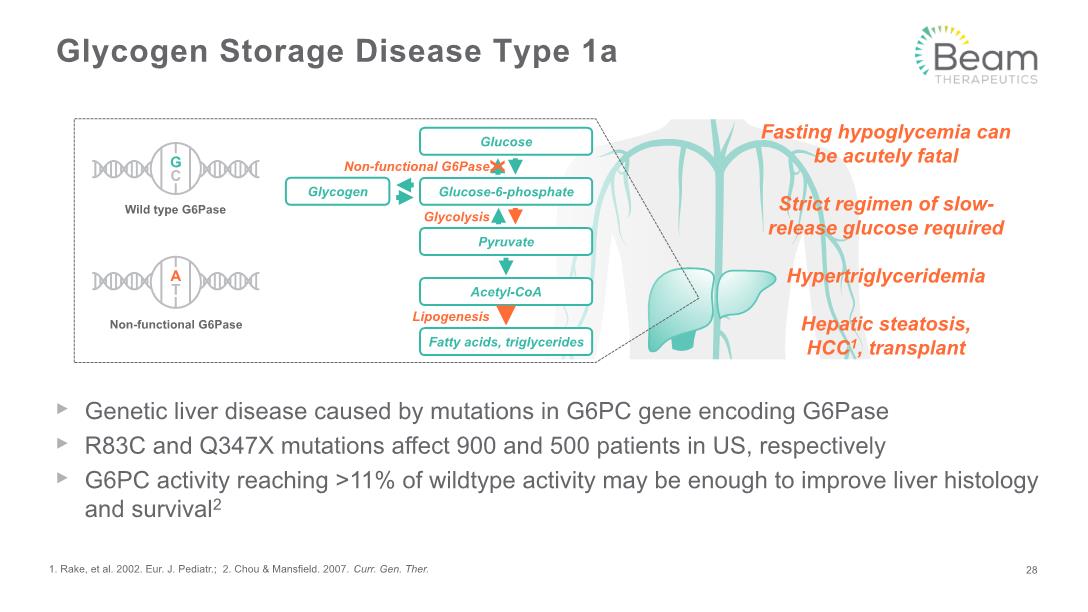
Glycogen Storage Disease Type 1a Genetic liver disease caused by mutations in G6PC gene encoding G6Pase R83C and Q347X mutations affect 900 and 500 patients in US, respectively G6PC activity reaching >11% of wildtype activity may be enough to improve liver histology and survival2 1. Rake, et al. 2002. Eur. J. Pediatr.; 2. Chou & Mansfield. 2007. Curr. Gen. Ther. 28 Non-functional G6Pase Glycolysis Lipogenesis Glucose Glucose-6-phosphate Pyruvate Acetyl-CoA Fatty acids, triglycerides Glycogen X Fasting hypoglycemia can be acutely fatal Strict regimen of slow-release glucose required Hypertriglyceridemia Hepatic steatosis, HCC1, transplant
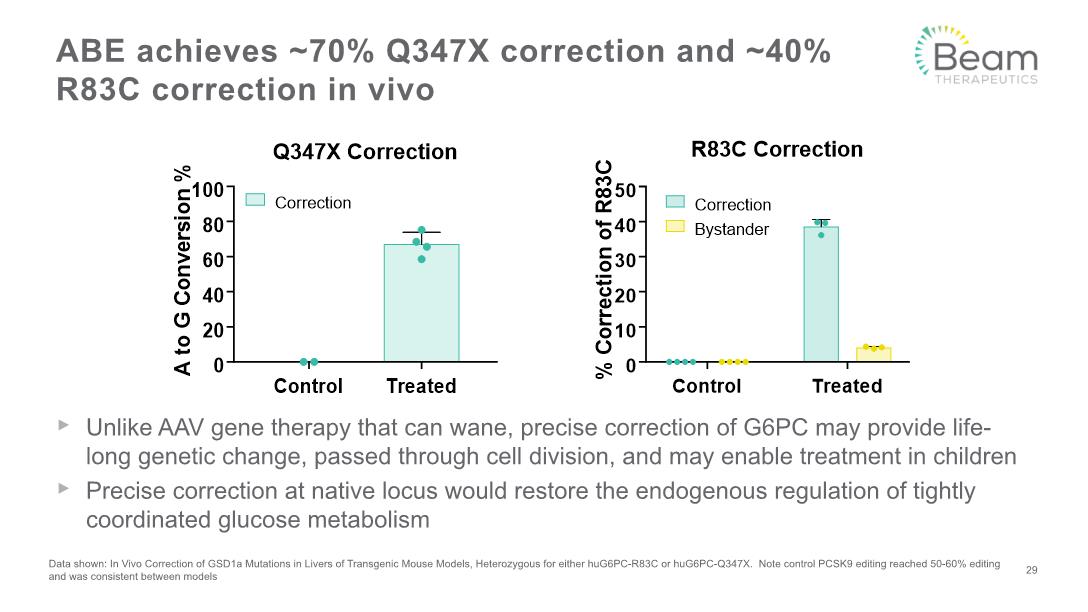
Unlike AAV gene therapy that can wane, precise correction of G6PC may provide life-long genetic change, passed through cell division, and may enable treatment in children Precise correction at native locus would restore the endogenous regulation of tightly coordinated glucose metabolism ABE achieves ~70% Q347X correction and ~40% R83C correction in vivo Data shown: In Vivo Correction of GSD1a Mutations in Livers of Transgenic Mouse Models, Heterozygous for either huG6PC-R83C or huG6PC-Q347X. Note control PCSK9 editing reached 50-60% editing and was consistent between models 29

Clinically validated technology for in vivo delivery to a variety of tissue types Split intein technology designed to deliver base editor and guide RNA by co-infection with two viruses DNA base editors are larger than the ~4.5kb limit of AAV vectors Viral delivery for in vivo base editing 30 Co-infection Split editor AAV 1 2 1st half of editor 2nd half of editor and gRNA Full-length editor protein 3 +

ABCA4 mutations cause central vision loss and death of photoreceptor cells Most prevalent G1961E mutation accounts for 5,500 patients in US No approved therapies today ABCA4 gene cannot be packaged into a single AAV vector due to size Stargardt Macular Dystrophy ABE achieves ~75% correction delivered with split AAV 31 Retina Image Bank Editing levels in excess of 12-20% expected to be disease-modifying Data shown: ABE editing of *surrogate base after split AAV delivery in retinal cells (N=3). *Surrogate base is the third base in the disease target GAA codon, which is also present in normal non-Stargardt cells; MOI = Multiplicity of Infection Weeks:

Strategic collaborations and investment in future capabilities 32 Base editing for the prevention of cardiovascular disease Non-ablative conditioning for improved hematology transplant in combination with BEAM-101 and BEAM-102 Clinical and research activities for sickle cell disease, leukemias, and other diseases Preclinical research collaboration for base editing in Stargardt Disease Custom build of 100,000 ft2 cGMP facility supporting ex vivo and in vivo clinical and commercial manufacturing for base editing programs Developing potential best-in-class disease-specific base editing therapies Translational and clinical research with world-class teams Critical internal capabilities to accelerate future development

Meet the Beam Team John Evans Chief Executive Officer Giuseppe Ciaramella, PhD President, Chief Scientific Officer Courtney Wallace Chief Business Officer Susan O’Connor Chief Human Resources Officer Christine Bellon PhD, JD SVP, Chief Legal Officer Suzanne Fleming SVP, Finance Terry-Ann Burrell Chief Financial Officer Collectively contributed to development of 13 approved products and 70 IND filings Brian Riley SVP, Technical Operations 33

Thank you 34
